20110606-fix-update_compositions-swissmedic_plugin-testcases-feedbacks-writer-assign_deprived_sequence-ch_oddb
<< | Index | >>
- Goal/Estimate/Evaluation
- Fix update_sequence / 80% / 100%
- Milestones
Check wrong points- Fix product name
- Fix active agents
- Summary
- Commits
- Fixed swissmedic plugin to update product name and active agents correctly (ch.oddb)
- Updated testcases, ext/chapterparse/test/test_writer.rb, view/admin/assign_deprived_sequence.rb, view/drugs/feedbacks.rb (ch.oddb)
Check the wrong points
- Status: Masa fix-active_agents-testcases-view-activeagent-suggest_address-narcotics-ch_oddb
- Sérocytol, Iscador
Goal
- Product name:
- scan backwards from the product name until the first comma and cut everything off until the first comma, looking at the string from the end.
- So in my case "Injektionslösung" would be cut off as that does not belong into the name.
- Active agents:
- the active agents of Sérocytol are not recognized correctly
- the column O of Packungen.xls has active agents with values in the brackets
- the method, fix_compositions, separates the values in the brackets into individual active agents
Fix product name
Check src/plugin/swissmedic.rb#update_sequence
def update_sequence(registration, row, opts={:create_only => false})
seqnr = "%02i" % cell(row, column(:seqnr)).to_i
ptr = if(sequence = registration.sequence(seqnr))
return sequence if opts[:create_only]
sequence.pointer
else
(registration.pointer + [:sequence, seqnr]).creator
end
## some names use commas for dosage
parts = cell(row, column(:name_base)).split(/\s*,(?!\d|[^(]+\))\s*/u)
descr = parts.pop
## some names have dosage data after the galenic form
if /[\d\s][m]?[glL]\b/.match(descr) && parts.size > 1
descr = parts.pop << ', ' << descr
end
base = parts.join(', ')
base, descr = descr, nil if base.empty?
...
args = {
:composition_text => ctext,
:name_base => base,
:name_descr => descr,
:dose => nil,
:sequence_date => date_cell(row, column(:sequence_date)),
:export_flag => nil,
}
Test
namestr = "Iscador M 0,0001 mg, Injektionslösung, anthroposophisches Arzneimittel"
parts = namestr.split(/\s*,(?!\d|[^(]+\))\s*/u)
descr = parts.pop
print "parts = "
p parts
print "descr = "
p descr
if /[\d\s][m]?[glL]\b/.match(descr) && parts.size > 1
descr = parts.pop << ', ' << descr
end
base = parts.join(', ')
puts
print "parts = "
p parts
print "base = "
p base
print "descr = "
p descr
Result
parts = ["Iscador M 0,0001 mg", "Injektionsl\303\266sung"] descr = "anthroposophisches Arzneimittel" parts = ["Iscador M 0,0001 mg", "Injektionsl\303\266sung"] base = "Iscador M 0,0001 mg, Injektionsl\303\266sung" descr = "anthroposophisches Arzneimittel"
Note
- /\s*,(?!\d|[^(]+\))\s*/u means
- basically, the string is split by 'comma(,)' but if there is a digit (\d) or a blaket ('(') after the comma then it does not split the string
Consideration
- The algorithm to take name_base and name_descr is as follows:
- split the data of column C 'Sequenzname' by comma
- basically, the last element will be the name_descr and the rest parts will be the name_base
Re-design the algorithm
- the name_base (Präparatname) should be the first element parts that are split the column C data by commma
- the name_descr (Beschreibung) should be the rest parts
Experiment (src/plugin/swissmedic.rb#update_sequence)
def update_sequence(registration, row, opts={:create_only => false})
seqnr = "%02i" % cell(row, column(:seqnr)).to_i
ptr = if(sequence = registration.sequence(seqnr))
return sequence if opts[:create_only]
sequence.pointer
else
(registration.pointer + [:sequence, seqnr]).creator
end
## some names use commas for dosage
parts = cell(row, column(:name_base)).split(/\s*,(?!\d|[^(]+\))\s*/u)
base = parts.shift
# descr = parts.pop
## some names have dosage data after the galenic form
# if /[\d\s][m]?[glL]\b/.match(descr) && parts.size > 1
# descr = parts.pop << ', ' << descr
# end
# base = parts.join(', ')
# base, descr = descr, nil if base.empty?
descr = unless parts.empty?
parts.join(', ')
else
nil
end
Run
- fix_compositions
masa@masa ~/ywesee/oddb.org $ bin/admin ch.oddb> ODDB::SwissmedicPlugin.new(self).fix_compositions
Result
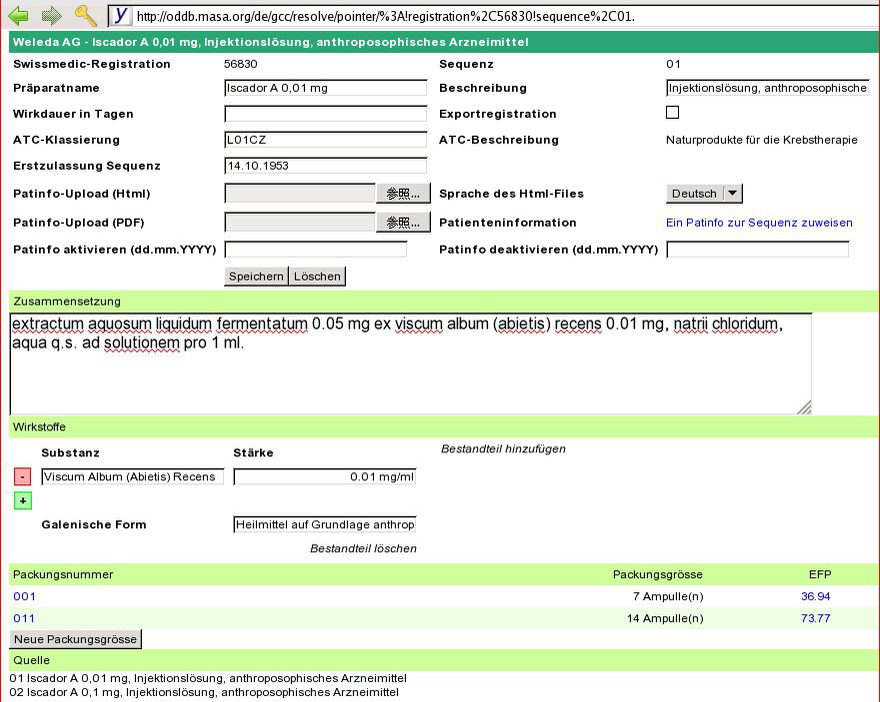
Fix active agents
Problem
- The data inside brackets of column O data is split by commma
Solution
- the comma inside brackets should be ignored for the splitting
Check
def update_compositions(seq, row, opts={:create_only => false})
if opts[:create_only] && !seq.active_agents.empty?
seq.compositions
elsif(namestr = cell(row, column(:substances)))
res = []
names = namestr.split(/\s*,\s*/u).collect { |name|
capitalize(name) }.uniq
substances = names.collect { |name|
update_substance(name)
}
...
Test
namestr = 'globulina equina (Immunisation mit Schweine-Knochen, -Knorpel, -Bindegewebe, -Serosa, -Lymphknoten)' parts = namestr.split(/\s*,(?!\d|[^(]+\))\s*/u) p parts namestr = 'viscum album (mali) recens, argenti carbonas (0,01 ug pro 100 mg herba recente)' parts = namestr.split(/\s*,(?!\d|[^(]+\))\s*/u) p parts
Result
["globulina equina (Immunisation mit Schweine-Knochen, -Knorpel, -Bindegewebe, -Serosa, -Lymphknoten)"] ["viscum album (mali) recens", "argenti carbonas (0,01 ug pro 100 mg herba recente)"]
Experiment (src/plugin/swissmedic.rb#update_compositions)
def update_compositions(seq, row, opts={:create_only => false})
if opts[:create_only] && !seq.active_agents.empty?
seq.compositions
elsif(namestr = cell(row, column(:substances)))
res = []
#names = namestr.split(/\s*,\s*/u).collect { |name|
names = namestr.split(/\s*,(?!\d|[^(]+\))\s*/u).collect { |name|
capitalize(name) }.uniq
Run
- fix_compositions
masa@masa ~/ywesee/oddb.org $ bin/admin ch.oddb> ODDB::SwissmedicPlugin.new(self).fix_compositions
Result
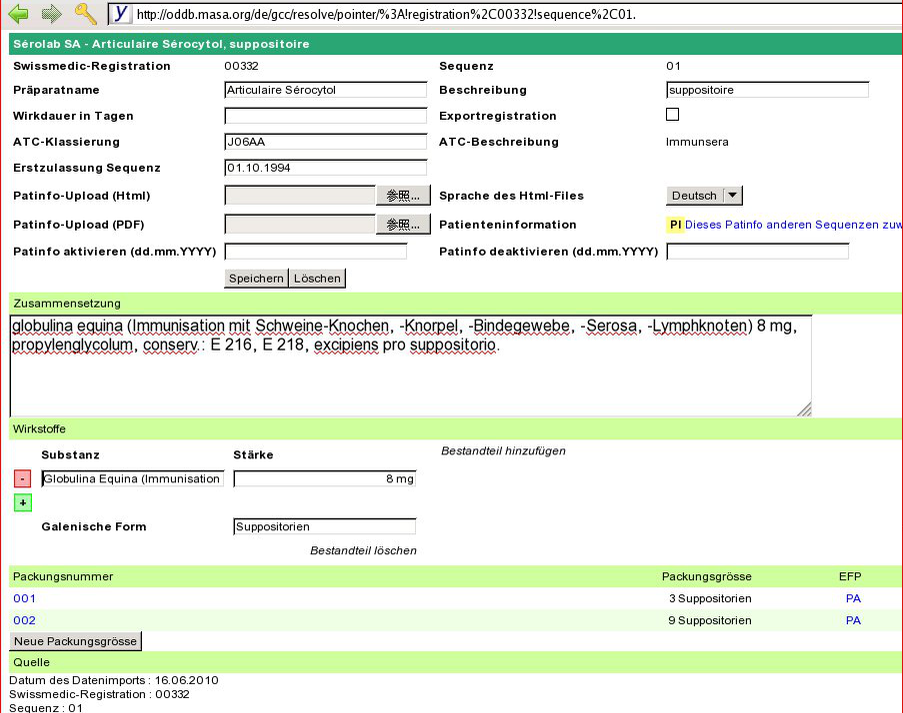
Commit
Testcases ch.oddb
src/view/drugs/feedbacks.rb (coverage: 100%)ext/chapterparse/src/writer.rb (coverage: 100%)src/view/admin/assign_deprived_sequence.rb (coverage: 100%)- src/view/drugs/resultlimit.rb (coverage: 74.00%)
- src/plugin/analysis.rb (coverage: 20.20%)
Commit