20130716-import-company
<< | Index | >>
Summary
- Import Company imports wrong FI
Commits
- https://github.com/ngiger/oddb.org/commit/a296e4d6f4162a58572def4b14385b185f108d04 Fix import swissmedic by companies
Index
FI without correct line breaks
Why have the following FI have too long lines?
- http://ch.oddb.org/de/gcc/fachinfo/swissmedicnr/51759
- http://ch.oddb.org/de/gcc/fachinfo/swissmedicnr/51608
I consider this problem was resolved when I fixed the problem with the missing spaces before the Wirkstoff, as there was an inbalanced </span> present. After running the import on my own VM the page displayed correctly, see 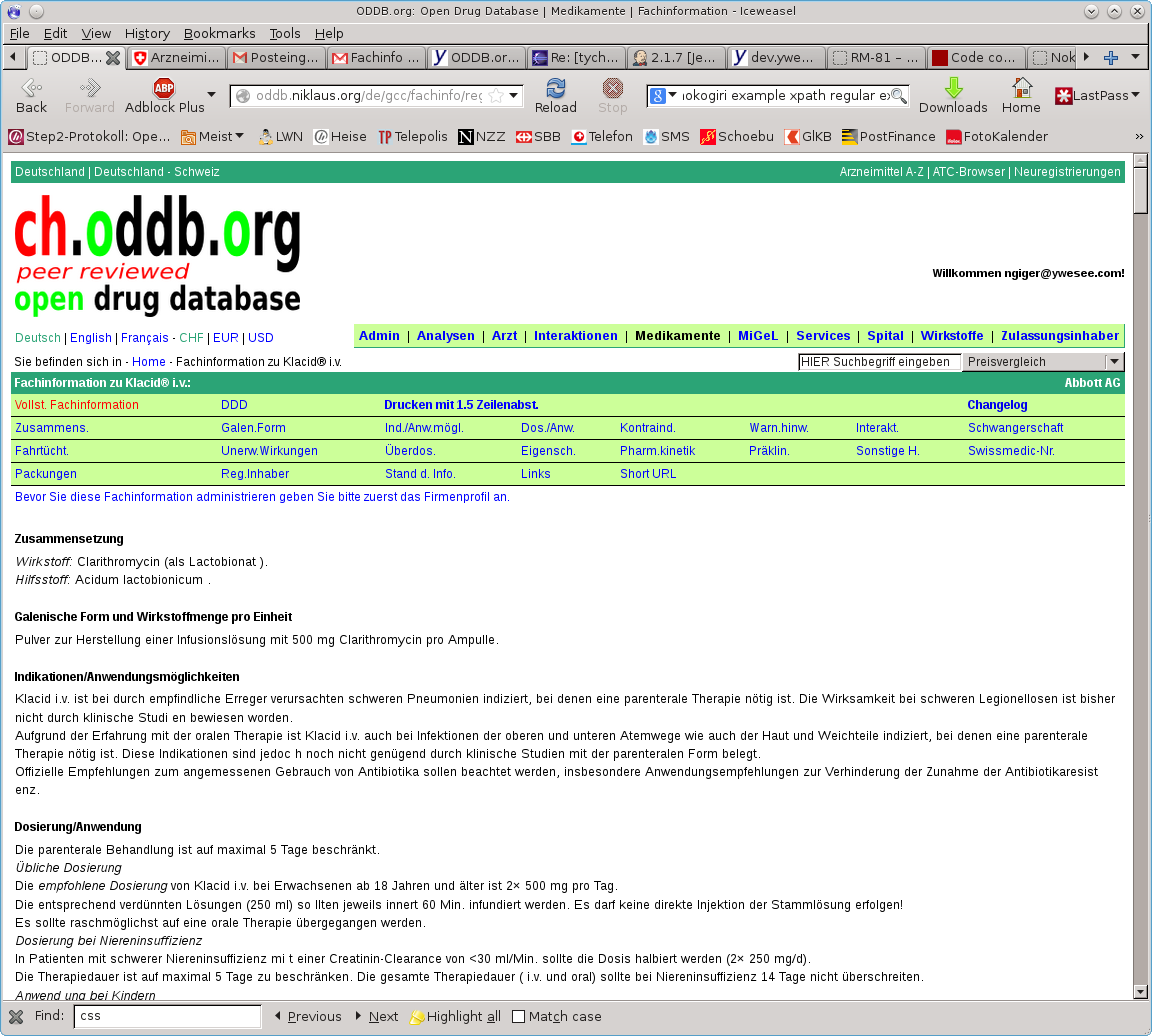
Import Company imports wrong FI
Running an import for jobs/update_textinfo_swissmedicinfo no-download --target=fi --reparse Crucell gives the following output
textinfo_swissmedicinfo_company_index returns 467,704. See /var/www/oddb.org/data/html/Crucell_index.html
import_swissmedicinfo_by_iksnrs [467, 704]
import_swissmedicinfo_by_iksnrs iksnr 467 {}
parse_and_update: calls parse_fachinfo, /var/www/oddb.org/data/html/fachinfo/de/Accupaque__swissmedicinfo.html, name Accupaque™ de title Accupaque™, styles p{margin-top:0pt;margin-right:0pt;margin-bottom:0pt;
Looking at the XML-file wie expect the following IDs
00467 1 Vivotif, Kapseln CRUCELL SWITZERLAND AG 08.08. J07AP01 00572 1 Epaxal, Injektionssuspension CRUCELL SWITZERLAND AG 08.08. J07BC02 00614 12 Inflexal V, Injektionssuspension CRUCELL SWITZERLAND AG 08.08. J07BB02 00668 1 Gamunex 10%, Lösung zur intravenösen Injektion CRUCELL SWITZERLAND AG 08.09. J06BA02 00697 1 Prolastin, Trockensubstanz zur Herstellung einer Infusionslösung CRUCELL SWITZERLAND AG 06.01.1. B02AB02 00704 2 Dukoral, orale Suspension CRUCELL SWITZERLAND AG 08.08. J07AE01
- Found that you must pass to update_textinfo_swissmedicinfo a five digit number 00467 if you want to import iksnr 467.
- I changed the code to use
sprintf("%05d", id)instead ofid - Found that we always search for Patienteninfo, even when Fachinfo was specified.
Still having the problem that calling jobs/update_textinfo_swissmedicinfo no-download --target=both --reparse 467 returns wrong patinfo 56079
import_swissmedicinfo_by_iksnrs ["00467"] target both
import_swissmedicinfo_by_iksnrs iksnr "00467" {}
parse_and_update: calls parse_fachinfo, /var/www/oddb.org/data/html/fachinfo/de/Vivotif__swissmedicinfo.html, name Vivotif® de title Vivotif®, styles p{margin-top:0pt;margin-right:0pt;margin-bottom:0pt;margin-left:0pt;
parse_and_update: update_fachinfo name Vivotif® returned ["00467"]
parse_and_update: calls parse_fachinfo, /var/www/oddb.org/data/html/fachinfo/fr/Vivotif__swissmedicinfo.html, name Vivotif® fr title Vivotif®, styles p{margin-top:0pt;margin-right:0pt;margin-bottom:0pt;margin-left:0pt;
parse_and_update: update_fachinfo name Vivotif® returned ["00467"]
parse_and_update: calls parse_patinfo, /var/www/oddb.org/data/html/patinfo/de/Arcoxia__swissmedicinfo.html, name Arcoxia® de title Arcoxia®, styles p{margin-top:0pt;margin-right:0pt;margin-bottom:0pt;margin-left:0pt;
parse_and_update: update_patinfo name Arcoxia® returned ["56079"]
parse_and_update: calls parse_patinfo, /var/www/oddb.org/data/html/patinfo/fr/Arcoxia__swissmedicinfo.html, name Arcoxia® fr title Arcoxia®, styles p{margin-top:0pt;margin-right:0pt;margin-bottom:0pt;margin-left:0pt;
parse_and_update: update_patinfo name Arcoxia® returned ["56079"]
Wrote a simple test script to show that the procedure extract_matched_name in text_info.rb does not work correctly. There are no test-cases present.
Added the test to test/test_plugin/text_info.rb.
The import of Bayer seems to be correct now, but I got another time the error @|@Message: ERROR: prepared statement "ruby-dbi:Pg:5950206401373984273.0981805" already exists@@
Pushed the commit Fix import swissmedic by companies which make the following changes
- Use XML-element authNrs to get the iksnr (old solution parsed HTML of the XML-element content)
- Handle cases where comma separated iksnr were used in the XML
Following curious situation found. There are 4 iksnrs in the XML consisting of a single '-', e.g for Sérocytol®. On the swissmedic-HTML you find 00332, 00328, 00278, 00279, 00277, 00282, 00284, 00286, 00288, 00294, 00295, 00296, 00300, 00306, 00309, 00313, 60026, 00314, 00315, 00316, 00299, 00330, 00334, 00335, 00338, 00339 (Swissmedic).
It might be more consistent to get the iksnr of each company using also the AipsDownload_latest.xml instead of using mechanize to get the information from http://www.swissmedicinfo.ch.