20170906-ethereum-contract
Summary
- Ethereum contracs
- Update_drugshortage did not create data/downloads/drugshortage.csv
- Allow users to display only selected SL_categories
- Problem converting oddb2xml oddb_with_migel.dat to ISO-8859-1
- Keep in Mind
Commits
- https://github.com/ngiger/oddb.org/commit/59752467bb7b6034d6e2ff621183f3014cf8b73c Create downloads/drugshortage.csv if missing
- https://github.com/ngiger/oddb.org/commit/66652d942479316ae2ab7b3c0c896a97a27c2355 Ident with space
- https://github.com/ngiger/oddb.org/commit/a8f21b6ee1e35408444d3b0d6735da111d4feaf8 Adding preferences to select only SL, SL-Types, valid drugs. 23
- https://github.com/ngiger/oddb2xml/commit/47023837299a18d1fe698eca52ff5691002c95fe Before emitting lines, check for UTF-8
- https://github.com/ngiger/oddb2xml/commit/575a5a6abd58a00715222a9785f7eccb3be945e3 Bumped version to 2.4.2
Index
- Ethereum contracs
- Update_drugshortage did not create data/downloads/drugshortage.csv
- Allow users to display only selected SL_categories
- Problem converting oddb2xml oddb_with_migel.dat to ISO-8859-1
Update_drugshortage did not create data/downloads/drugshortage.csv
Remarked that yesterday jobs/update_drugshortage did not create data/downloads/drugshortage.csv on thinpower. Also yesterday CSV which was identical to today was not removed. Fixed with commit Create downloads/drugshortage.csv if missing
Now I must somehow persist the changes. Probably similar to the ZSR-ID for which we have a javascript under doc/resources/javascript/prescription.js.
To limit the search result I tried to add the following lines in the method _search_drugs_state of src/state/global.rb
if @session.user_input(:search_imitation_SL_only) || true
puts "Before filtering for search_imitation_SL_only we have #{result.package_count} results"
result.each{|atc| atc.packages.reject! { |pack| !pack.sl_entry }}
count = 0; result.each{|atc| atc.packages.each { |pack| count+=1 }}; count
puts "After filtering we have #{count} packages in result"
end
This displays correctly only 10 items for interal but still says Propranolol (C07AA05) 11 Präparate. And in in the log I get the following error
Before filtering for search_imitation_SL_only we have 11 results NoMethodError undefined method `user' for nil:NilClass /var/www/oddb.org/src/util/resultsort.rb:133:in `adjusted_name_and_prio' /var/www/oddb.org/src/util/resultsort.rb:75:in `block in sort_result' /var/www/oddb.org/src/util/resultsort.rb:74:in `each' /var/www/oddb.org/src/util/resultsort.rb:74:in `sort_by' /var/www/oddb.org/src/util/resultsort.rb:74:in `sort_by!' /var/www/oddb.org/src/util/resultsort.rb:74:in `sort_result' /var/www/oddb.org/src/model/search_result.rb:49:in `packages' /var/www/oddb.org/src/state/global.rb:930:in `block in _search_drugs_state' /var/www/oddb.org/src/model/search_result.rb:131:in `each' /var/www/oddb.org/src/model/search_result.rb:131:in `each' /var/www/oddb.org/src/state/global.rb:930:in `_search_drugs_state' /var/www/oddb.org/src/state/global.rb:873:in `search' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/state.rb:198:in `_trigger' /var/www/oddb.org/src/state/global.rb:1051:in `_trigger' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/state.rb:187:in `trigger' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/session.rb:256:in `block in process_rack' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/session.rb:209:in `synchronize' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/session.rb:209:in `process_rack' /var/www/oddb.org/vendor/ruby/2.4.0/gems/sbsm-1.5.9/lib/sbsm/app.rb:127:in `call' <...> After filtering we have 10 packages in result
This is probably not the correct approach. NO. It could be fixed by two checks for nil in the util/result_sort.rb. Saved my work as ranch search_prefs
The following problems need to be resolved next week:
- Correct package_count after filtering the result (or improving the model/search_result with a filter)
- Persist filtering for SL-entry/types/only valid packages
Ethereum contracs
For the ruby gem ethereum.rb I must install | prerequisites, which needs a tool called parity. Downloaded https://github.com/paritytech/parity/archive/v1.7.0.tar.gz to /usr/local/src
bash <(curl https://get.parity.io -Lk) sudo npm install -g solc
Allow users to display only selected SL_categories
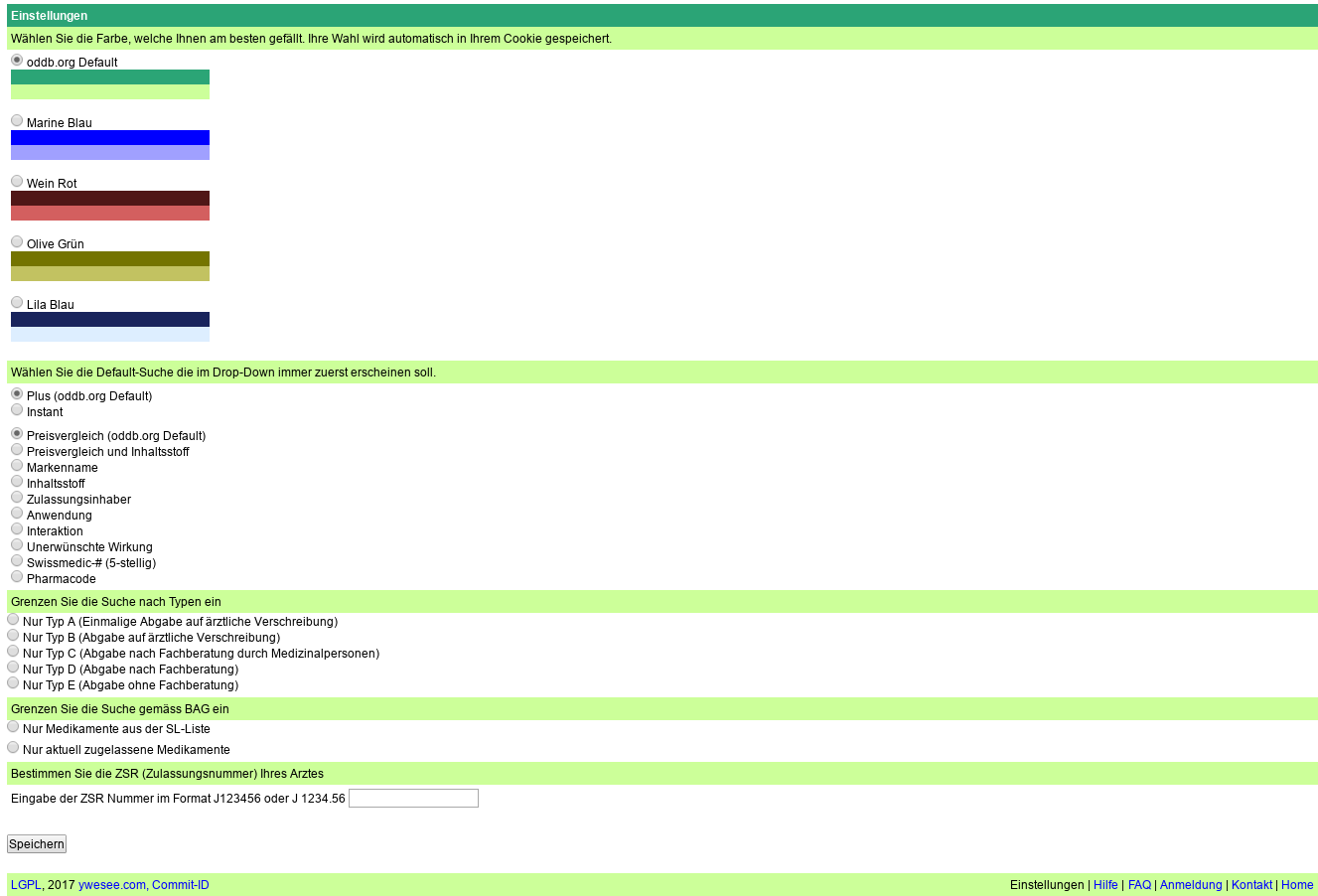
In the configuration of ch.oddb.org we want to allow the users to opt-out of displaying individual SL-Categories and pending registrations. By default all categories and pending registrations are displayed.
Adding new texts in src/custom/lookandfeelbase.rb (after converting all tabs to spaces).
Added also a sub-heading before the ZSR. Now my screen looks like this
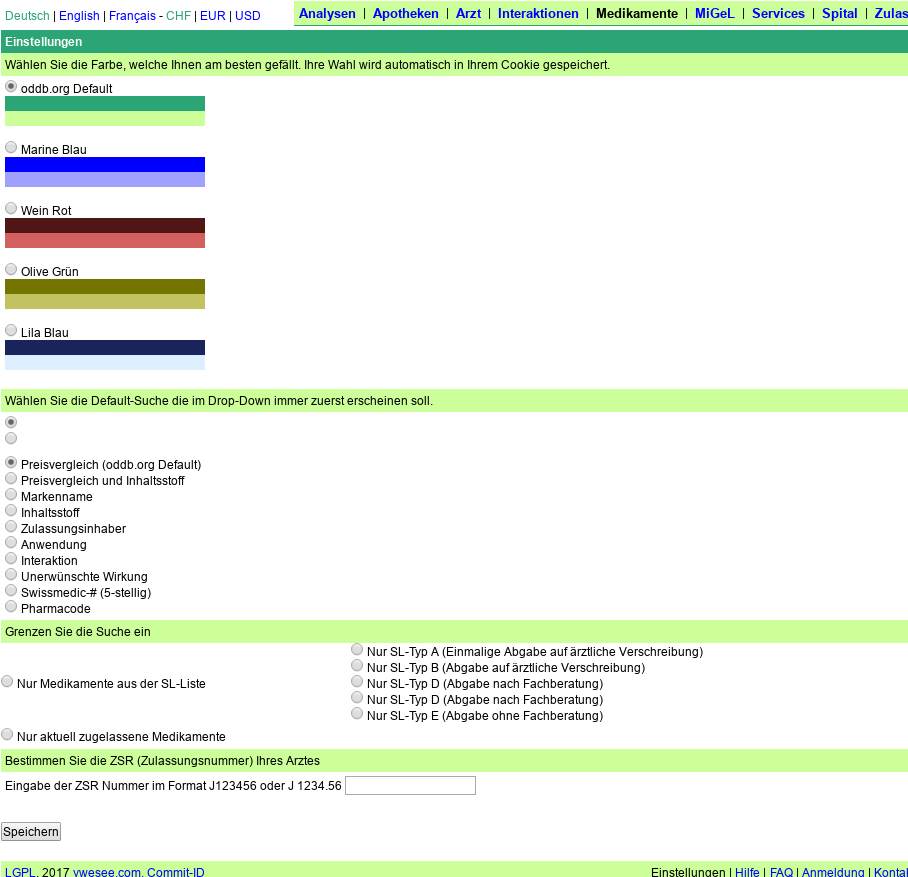
After some corrections
Problem converting oddb2xml oddb_with_migel.dat to ISO-8859-1
Somehow iconv cannot convert the UTF-8 file to 88519 as seen by this snippet
iconv -f utf-8 -t ISO-8859-1 -o oddb_with_migel.dat.8559 oddb_with_migel.dat; file oddb_with_migel.dat.8559 oddb_with_migel.dat.8559: UTF-8 Unicode text, with CRLF line terminators ls -lrt oddb_with_migel.* -rw-r--r-- 1 niklaus users 4574742 Sep 6 10:36 oddb_with_migel.dat -rw-r--r-- 1 niklaus users 4564763 Sep 6 11:46 oddb_with_migel.dat.8559 diff oddb_with_migel.dat oddb_with_migel.dat.8559 | head 1c1 < 1110361815SEROCYTOL Herz-Gefässe Supp 3 Stk 0000000000003030000000076800027700142 --- > 1110361815SEROCYTOL Herz-Gefässe Supp 3 Stk 0000000000003030000000076800027700142 16,17c16,17 < 1120658018SEROCYTOL Leber-Gefässe-Rektum Supp 3 Stk 0022840035203030000000076800030000112 < 1121550754SEROCYTOL Leber-Gefässe-Rektum Supp 9 Stk 0056930076903030000000076800030000282 --- > 1120658018SEROCYTOL Leber-Gefässe-Rektum Supp 3 Stk 0022840035203030000000076800030000112 > 1121550754SEROCYTOL Leber-Gefässe-Rektum Supp 9 Stk 0056930076903030000000076800030000282
Must run oddb2xml -a -f dat and create the oddb_with_migel.dat with encoding latin.
I cannot use latin or latin-1 as ruby (at least 2.4.1) says /home/niklaus/git/oddb2xml/lib/oddb2xml/cli.rb:139: warning: Unsupported encoding latin ignored
Script is now
#!/usr/bin/env ruby
input = ARGV[0]
content = IO.read(input)
puts "converting utf-8 #{input} #{content.encoding} -> ISO-8859-1"
File.open(input, 'w+:ISO-8859-1') { |f| f.write(content.encode('ISO-8859-1'))}
file_content = File.open(input, &:read)
puts "encoding is now #{file_content.encoding}"
Gives the error
converting utf-8 oddb_with_migel.dat UTF-8 -> ISO-8859-1
./utf8_to_8859.rb:6:in `encode': U+2013 from UTF-8 to ISO-8859-1 (Encoding::UndefinedConversionError)
from ./utf8_to_8859.rb:6:in `block in <main>'
from ./utf8_to_8859.rb:6:in `open'
from ./utf8_to_8859.rb:6:in `<main>'
We have several lines that give problems. At the moment I just ignore them with this variant of my script
#!/usr/bin/env ruby
input = ARGV[0]
content = IO.read(input)
lines = IO.readlines(input)
puts "converting utf-8 #{input} #{content.encoding} -> ISO-8859-1"
File.open(input, 'w+:ISO-8859-1') do
|f|
lines.each do |line|
begin
f.write(line.encode('ISO-8859-1'))
rescue => error
puts "#{error}: Ignoring #{line}"
end
end
end
file_content = File.open(input, &:read)
puts "encoding is now #{file_content.encoding}"
This gives the followin output
U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116346515SENSURA Mio 1t Uro 10–33 midi con lig so op 10 Stk000000000000300 000000057089326174201 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116346521SENSURA Mio 1t Uro 10–33 maxi con lig so op 10 Stk000000000000300 000000057089326174511 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116267081SENSURA Mio 1t Uro 10–43 maxi conv trans 10 Stk 000000000000300 000000057089328855221 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116267075SENSURA Mio 1t Uro 10–23 maxi conv trans 10 Stk 000000000000300 000000057089328855461 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266992SENSURA Mio 1t Uro 10–23 maxi conv opaq 10 Stk 000000000000300 000000057089328856141 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266934SENSURA Mio 1t Uro 10–43 maxi conv lig tran 10 Stk000000000000300 000000057089328856521 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266928SENSURA Mio 1t Uro 10–23 maxi conv lig tran 10 Stk000000000000300 000000057089328856761 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266851SENSURA Mio 1t Uro 10–23 maxi conv lig opaq 10 Stk000000000000300 000000057089328857441 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266822SENSURA Mio 1t Uro 10–23 midi conv lig opaq 10 Stk000000000000300 000000057089328857751 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266816SENSURA Mio 1t Uro 10–50 ma con lig sof tra 10 Stk000000000000300 000000057089328857821 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266727SENSURA Mio 1t off opa 10–23 Fens conv maxi 10 Stk000000000000300 000000057089328858501 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266644SENSURA Mio 1t off opa 10–23 Fen con lig mx 10 Stk000000000000300 000000057089328859281 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266615SENSURA Mio 1t off opa 10–23 Fen con lig md 10 Stk000000000000300 000000057089328859591 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266561SENSURA Mio 1t of op 10–50 Fe con lig so mx 10 Stk000000000000300 000000057089328860001 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266466SENSURA Mio 1t ges opa 10–23 Fen conv maxi 10 Stk 000000000000300 000000057089328860931 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266420SENSURA Mio 1t ges opa 10–23 Fen conv midi 10 Stk 000000000000300 000000057089328861231 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266348SENSURA Mio 1t ges opa 10–23 Fen con lig mx 10 Stk000000000000300 000000057089328861921 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266319SENSURA Mio 1t ges opa 10–23 Fen con lig md 10 Stk000000000000300 000000057089328862221 U+2013 from UTF-8 to ISO-8859-1: Ignoring 1116266302SENSURA Mio 1t ge op 10–50 Fe con lig so mx 10 Stk000000000000300 000000057089328862391 encoding is now UTF-8 > file oddb_with_migel.dat oddb_with_migel.dat: ISO-8859 text
Converted the refdata_NonPharma.xml with xmllint into a nice format and found there
<ITEM DT="2017-09-06T00:00:00">
<ATYPE>NONPHARMA</ATYPE>
<GTIN>5708932617420</GTIN>
<PHAR>6346515</PHAR>
<NAME_DE>SENSURA Mio 1t Uro 10–33 midi con lig so op 10 Stk</NAME_DE>
<NAME_FR>SENSURA Mio 1t Uro 10–33 midi con lig so op 10 pce</NAME_FR>
<AUTH_HOLDER_NAME>Coloplast AG (Euro 1)</AUTH_HOLDER_NAME>
<AUTH_HOLDER_GLN>7601001370494</AUTH_HOLDER_GLN>
</ITEM>
No I must modify the source to catch read/writing this error. But first I will to it my small script.
Even better would be outputting a message and replacing the wrong characters with a '?'. Scripts looks now like this
#!/usr/bin/env ruby
input = ARGV[0]
content = IO.read(input)
lines = IO.readlines(input)
puts "converting utf-8 #{input} #{lines.size} lines #{content.encoding} -> ISO-8859-1"
File.open(input, 'w+:ISO-8859-1') do
|f|
lines.each do |line|
begin
f.write(line.encode('ISO-8859-1'))
rescue => error
puts "#{error} #{line}"
f.write(line.encode('ISO-8859-1', :replace => '?'))
end
end
end
and produces
> ./utf8_to_8859.rb oddb_with_migel.dat converting utf-8 oddb_with_migel.dat 46058 lines UTF-8 -> ISO-8859-1 U+2013 from UTF-8 to ISO-8859-1 1116346515SENSURA Mio 1t Uro 10–33 midi con lig so op 10 Stk000000000000300 000000057089326174201 U+2013 from UTF-8 to ISO-8859-1 1116346521SENSURA Mio 1t Uro 10–33 maxi con lig so op 10 Stk000000000000300 000000057089326174511 U+2013 from UTF-8 to ISO-8859-1 1116267081SENSURA Mio 1t Uro 10–43 maxi conv trans 10 Stk 000000000000300 000000057089328855221 U+2013 from UTF-8 to ISO-8859-1 1116267075SENSURA Mio 1t Uro 10–23 maxi conv trans 10 Stk 000000000000300 000000057089328855461 U+2013 from UTF-8 to ISO-8859-1 1116266992SENSURA Mio 1t Uro 10–23 maxi conv opaq 10 Stk 000000000000300 000000057089328856141 U+2013 from UTF-8 to ISO-8859-1 1116266934SENSURA Mio 1t Uro 10–43 maxi conv lig tran 10 Stk000000000000300 000000057089328856521 U+2013 from UTF-8 to ISO-8859-1 1116266928SENSURA Mio 1t Uro 10–23 maxi conv lig tran 10 Stk000000000000300 000000057089328856761 U+2013 from UTF-8 to ISO-8859-1 1116266851SENSURA Mio 1t Uro 10–23 maxi conv lig opaq 10 Stk000000000000300 000000057089328857441 U+2013 from UTF-8 to ISO-8859-1 1116266822SENSURA Mio 1t Uro 10–23 midi conv lig opaq 10 Stk000000000000300 000000057089328857751 U+2013 from UTF-8 to ISO-8859-1 1116266816SENSURA Mio 1t Uro 10–50 ma con lig sof tra 10 Stk000000000000300 000000057089328857821 U+2013 from UTF-8 to ISO-8859-1 1116266727SENSURA Mio 1t off opa 10–23 Fens conv maxi 10 Stk000000000000300 000000057089328858501 U+2013 from UTF-8 to ISO-8859-1 1116266644SENSURA Mio 1t off opa 10–23 Fen con lig mx 10 Stk000000000000300 000000057089328859281 U+2013 from UTF-8 to ISO-8859-1 1116266615SENSURA Mio 1t off opa 10–23 Fen con lig md 10 Stk000000000000300 000000057089328859591 U+2013 from UTF-8 to ISO-8859-1 1116266561SENSURA Mio 1t of op 10–50 Fe con lig so mx 10 Stk000000000000300 000000057089328860001 U+2013 from UTF-8 to ISO-8859-1 1116266466SENSURA Mio 1t ges opa 10–23 Fen conv maxi 10 Stk 000000000000300 000000057089328860931 U+2013 from UTF-8 to ISO-8859-1 1116266420SENSURA Mio 1t ges opa 10–23 Fen conv midi 10 Stk 000000000000300 000000057089328861231 U+2013 from UTF-8 to ISO-8859-1 1116266348SENSURA Mio 1t ges opa 10–23 Fen con lig mx 10 Stk000000000000300 000000057089328861921 U+2013 from UTF-8 to ISO-8859-1 1116266319SENSURA Mio 1t ges opa 10–23 Fen con lig md 10 Stk000000000000300 000000057089328862221 U+2013 from UTF-8 to ISO-8859-1 1116266302SENSURA Mio 1t ge op 10–50 Fe con lig so mx 10 Stk000000000000300 000000057089328862391 > grep 708932617420 oddb_with_migel.dat 1116346515SENSURA Mio 1t Uro 10?33 midi con lig so op 10 Stk000000000000300 000000057089326174201
Pushed the commits