20130522-debug-patinfo-name-company-update
<< | Index | >>
Summary
- Debug Patinfo
- Some PIs have strange title and company name in view.
Commits
Index
Debug Patinfo name and company name
Some Patinfo have wrong name and company name in show view.
- Floxal
FI is correct.
- name
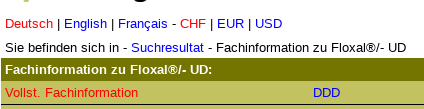
- company name
PI is wrong.
- name
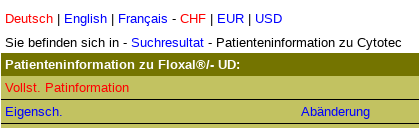
- company name

in :src/view/drugs/patinfo.rb:
class PatinfoPreviewComposite < HtmlGrid::Composite
COLSPAN_MAP = {
[0,1] => 2,
}
COMPONENTS = {
[0,0] => :patinfo_name,
[1,0] => :company,
[0,1] => View::Drugs::PatinfoInnerComposite,
}
...
end
There are strange relations via SequenceObserver.
# View [2] 1.9.3-p194(#<ODDB::View::Drugs::PatinfoComposite>)> model.class => ODDB::State::Drugs::Patinfo::PatinfoWrapper [3] 1.9.3-p194(#<ODDB::View::Drugs::PatinfoComposite>)> model.company_name => "Pfizer AG" # State [7] 1.9.3-p194(#<ODDB::State::Drugs::Patinfo>)> @model.class => ODDB::Patinfo [8] 1.9.3-p194(#<ODDB::State::Drugs::Patinfo>)> @model.sequences => [Cytotec, Tabletten, Fluconazol-Mepha 50 N, Kapseln, Fluconazol-Mepha 150 N, Kapseln, Fluconazol-Mepha 200 N, Kapseln, Amisulprid-Mepha 200, Tabletten, Amisulprid-Mepha 400, Lactab, Floxal, Augentropfen, Floxal UD, Augentropfen, Floxal, Augensalbe, Efudix, Salbe, Buscopan, Dragées, Buscopan, Suppositorien, Daivonex, Crème, Atorvastatin-Mepha 10 mg, Lactab, Atorvastatin-Mepha 20 mg, Lactab, Atorvastatin-Mepha 40 mg, Lactab, Atorvastatin-Mepha 80 mg, Lactab, Azithromycin-Mepha 250, Lactab, Azithromycin-Mepha 500, Lactab]
Some relations are wrong (Sequence and Patinfo).
- Atorvastin-Mepha(62211) ---> Floxal(51357, 51358, 55383) PI (wrong)
- Buscopan (17353) ---> Floxal(51357, 51358, 55383) PI (wrong)
- Fluconazol-Mepha (58063) ---> Floxal(51357, 51358, 55383) PI (wrong)
- Brufen (49669) has wrong sequence via SequencObserver
ch.oddb> patinfos.values.select{|pi| pi.sequences.is_a?(String) }.length
-> 1
db> patinfos.values.select{|pi| next if pi.sequences.is_a?(String); !pi.sequences.select{|seq| seq.patinfo != pi }.empty? }.length
-> 528
# Brufen
ch.oddb> patinfos.values.select{|pi| next if pi.sequences.is_a?(String); !pi.sequences.select{|seq| seq.patinfo != pi }.empty? }.first.name_base
-> Brufen
ch.oddb> patinfos.values.select{|pi| next if pi.sequences.is_a?(String); !pi.sequences.select{|seq| seq.patinfo != pi }.empty? }.first.sequences.length
-> 1
ch.oddb> patinfos.values.select{|pi| next if pi.sequences.is_a?(String); !pi.sequences.select{|seq| seq.patinfo != pi }.empty? }.first.sequences.first.patinfo.name_base
-> Alpicort-F
- PatinfoDocument Object is also correct.
- Some Patinfos (528 PIs) have strange Sequence (via SequenceObserver). (corrupted data ?)
Steps.
A. remove_sequence (Patinfo side)
- . Update
attr_readerof SequenceObserver.sequences toattr_accessor - . Reboot oddbd process
- . Replace a Invalid @sequences of PI (String one).
- . Back to attr_accessor to attr_reader.
- . Remove invalid Sequence connections (528 PIs)
- . Reboot
3.
ch.oddb> patinfos.values.select{|pi| if pi.sequences.is_a?(String) then pi.sequences = []; pi.sequences.odba_isolated_store; true; end }.length
-> 0
5. (do 4 times)
ch.oddb> patinfos.values.select {|pi| !pi.sequences.select {|seq| if seq.patinfo.odba_id != pi.odba_id then pi.remove_sequence(seq); true; end }.empty? }.length
-> xxx
# check count
ch.oddb> patinfos.values.select {|pi| !pi.sequences.select{|seq| seq.patinfo != pi }.empty? }.length
-> 0
B. reparse wrong patinfo (Sequence side)
There are strange connections also from Sequence to Patinfo.
ch.oddb> sequences.select {|seq| next unless seq.patinfo; !seq.patinfo.descriptions.values.select{|desc| next unless desc.iksnrs; desc.iksnrs.to_s.gsub(/[^0-9]/, '') !~ /#{seq.iksnr}/ }.empty? }.map(&:iksnr).uniq.length
-> 981
ch.oddb> sequences.select {|seq| next unless seq.patinfo; !seq.patinfo.descriptions.values.select{|desc| next unless desc.iksnrs; desc.iksnrs.to_s.gsub(/[^0-9]/, '') !~ /#{seq.iksnr}/ }.empty? }.first.iksnr
-> 59285
ch.oddb> sequences.select {|seq| next unless seq.patinfo; !seq.patinfo.descriptions.values.select{|desc| next unless desc.iksnrs; desc.iksnrs.to_s.gsub(/[^0-9]/, '') !~ /#{seq.iksnr}/ }.empty? }.first.patinfo.description("de").iksnrs
-> Zulassungsnummer
38219 (Swissmedic).
ch.oddb> sequences.select {|seq| next unless seq.patinfo; !seq.patinfo.descriptions.values.select{|desc| next unless desc.iksnrs; desc.iksnrs.to_s.gsub(/[^0-9]/, '') !~ /#{seq.iksnr}/ }.empty? }.first.patinfo.description("fr").iksnrs
-> Numéro d’autorisation
38219 (Swissmedic).
- commit
Attach: iksnrs.txt
Improve name detection of TextInfo Plugin.
- commit